Detecting Issues in Named Entity Recognition Datasets
In this tutorial, we’ll use Cleanlab Studio to automatically find label errors and other issues in Named Entity Recognition (NER) data. Label issues in NER datasets encompass problems like incorrect class labels, inconsistent choices across different data annotators, incorrect entity boundary labeling, ambiguity in entity types (multiple types might appear reasonable), entities overlooked by annotators, etc. Identifying and resolving these annotation problems and other data issues is essential for producing a high-quality dataset that can be used to train reliable NER models.

This tutorial considers the popular CONLL-2003 Named Entity Recognition dataset that contains labeled examples of entities in text classified into categories such as persons, organizations, and locations. You can replace this dataset with your own NER data as long as you transform it into the format described in the Download and Prepare Raw Dataset section.
(Note: this tutorial requires that you’ve created a Cleanlab account)
Install and import required dependencies
You can use pip to install all packages required for this tutorial as follows:
%pip install "cleanlab[datalab]" cleanlab-studio
from cleanlab_studio import Studio
from cleanlab.internal.token_classification_utils import get_sentence, filter_sentence, process_token, mapping, merge_probs
import os
from datasets import load_dataset
from ast import literal_eval
import numpy as np
import pandas as pd
pd.set_option('display.max_colwidth', None)
# you can find your Cleanlab Studio API key by going to studio.cleanlab.ai/upload,
# clicking "Upload via Python API", and copying the API key there
API_KEY = "<insert your API key>"
Download and Prepare Raw Dataset
We download the CONLL-2003 dataset into a data/ directory. This tutorial analyzes CONLL-2003 but you can use any dataset as long as it is either: stored in a similar format on your local machine as this CONLL-2003 dataset, or it is available via the Hugging Face datasets bank in a similar format to the TNER_huggingface dataset.
This tutorial focuses on using CONLL-2003 data which we download below. To run this notebook on your own entity recognition dataset, first convert it to the CONLL-2003 format.
wget -nc https://data.deepai.org/conll2003.zip && mkdir -p data
unzip conll2003.zip -d data/ && rm conll2003.zip
Accepted Local Format
This notebook will also run with a dataset that is stored locally. For local datasets, they need to be in a format similar to the CONLL-2003 data.
CONLL-2003 data are in the following format:
-DOCSTART- -X- -X- O
[word] [pos_tags] [chunk_tags] [ner_tags] ← Start of first sentence
...
[word] [pos_tags] [chunk_tags] [ner_tags]
[empty line]
[word] [pos_tags] [chunk_tags] [ner_tags] ← Start of second sentence
...
[word] [pos_tags] [chunk_tags] [ner_tags]
The ner_tags (named-entity recognition tags) are stored in the IOB2 format. CONLL-2003 includes the classes detailed below however custom ner_tags are also accepted.
ner_tags | Description |
|---|---|
O | Other (not a named entity) |
B-MIS | Beginning of a miscellaneous entity |
I-MIS | Miscellaneous entity |
B-PER | Beginning of a person entity |
I-PER | Person entity |
B-ORG | Beginning of an organization entity |
I-ORG | Organization entity |
B-LOC | Beginning of a location entity |
I-LOC | Location entity |
For more information, see here. For all local datasets, we cast all-caps words into lowercase except for the first character (eg. JAPAN -> Japan), to discourage the tokenizer from breaking such words into multiple subwords.
If you are working with data in similar format to CONLL-2003, set the dataset_path variable to either a single filepath or a list of all filepaths you want to upload to Cleanlab Studio.
Accepted Hugging Face format
Beyond running Cleanlab Studio on a locally-stored NER dataset in the CONLL-2003 format, you can also use a dataset stored in the Hugging Face datasets repository in TNER format. The TNER Organization hosts models and common datasets for T-NER, which is a python tool for language model fine-tuning on NER data. An example format of one dataset is below.
Here each example is made up of a list of tokens alongside a list of their respective named entity tags where for the i-th example, tokens[i][j] is the j-th token in the sentence with tag tags[i][j].
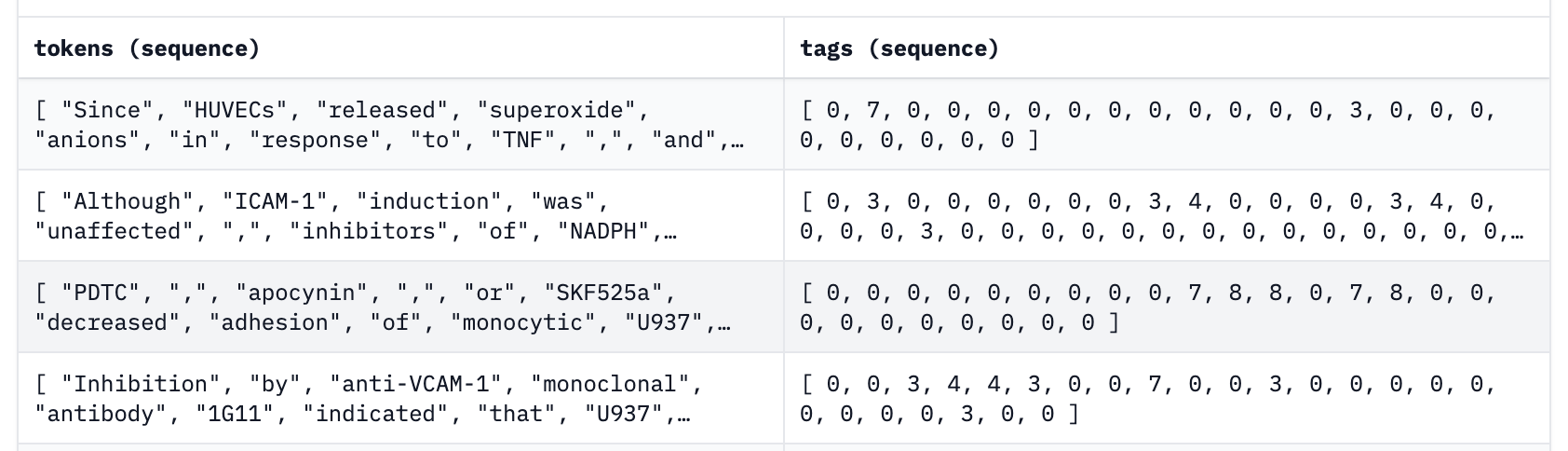
To upload your own dataset in TNER format, format each example into token and tag lists and then follow the instructions here then set the dataset_path variable to your dataset_repository/dataset_name or choose one of the datasets already provided by the organization.
Reformatting Named Entity Recognition Data
Occasionally, you may find improved performance in modifying the original Named Entities within a dataset before using Cleanlab Studio. This could involve consolidating ‘beginning’ and ‘continuation’ tags into a single named entity tag or eliminating specific word/tag combinations. To address this, we’ve introduced the reformat_ner_data() function below. This function accepts a list of filepaths in accepted local format and a dictionary that defines the desired transformations, and it generates new files in accepted format with the transformed tags.
To utilize this function, create a transformation_map dictionary. Each dictionary key represents a named entity in your dataset, and its corresponding value defines the desired transformation. When multiple keys share the same value, these entities will be merged in the resulting files. If a specific key has a value of ‘nan’, the corresponding tokens will be omitted from sentences in the new files.
Below is an example that demonstrates how to define transformations using the transformation_map for the CONLL-2003 example dataset.
transformation_map = {
"O": "nan",
"B-MISC": "nan",
"I-MISC": "nan",
"B-PER": "has_person",
"I-PER": "has_person",
"B-ORG": "has_organization",
"I-ORG": "has_organization",
"B-LOC": "has_location",
"I-LOC": "has_location",
}
This transfomation_map lists all the Named Entities found in the data as keys ['O', 'B-MISC', 'I-MISC', 'B-PER', 'I-PER', 'B-ORG', 'I-ORG', 'B-LOC', 'I-LOC']. The tokens marked as “O”, “B-MISC” and “I-MISC” are omitted from each sequence since these keys have nan set as their value. Entities marked as “B-PER” and “I-PER” are now marked as the same tag “has_person”. A similar union happens to “B-ORG” and “I-ORG”, which are united under “has_organization” and “B-LOC” and “I-LOC”, which become “has_location”. Basically we are saying our analysis only cares about the following entities: “person”, “organization”, “location”.
By passing this dictionary into the reformat_ner_data(filepath, transformation_map) function defined below, along with the desired filepath of the files you wish to transform, a new files featuring the updated entities will be generated. In the example below, you can locate the reformatted file at './data/transform_train.txt' or at a location defined by the optional parameter new_filepath.
filepath = './data/train.txt'
dataset_path = reformat_ner_data(filepath, transformation_map)
To reformat a dataset stored in the Hugging Face datasets repository, iterate through the examples and replace tags with their respective alternative in the transform_data_entity_map. Omit tags mapped to nan. Afterwards, upload the new dataset to Hugging Face and continue with the rest of the tutorial.
Optional: Define helper methods for formatting and analyzing NER data with Cleanlab
def reformat_ner_data(
filepath, transformed_filepath_dic, new_filepath=None, overwrite=False, sep=" "
):
"""Reformats data stored locally in the CONLL-2003 format using `transformed_filepath_dic`. Creates
new data files with applied transformations and returns their names. If overwrite is set to True
and new_filepath points to an already existing file, then data files are overwritten instead of
created.
Parameters
----------
filepath: str
Path pointing to local file in CONLL-2003 format.
transformed_filepath_dic: dict
Transformation dictionary where each key represents a named entity in your dataset, and its corresponding
value is a str of the desired transformation. If value is "nan", tokens with key are not included in the new file.
new_filepath: str
New filepath which the transformed data will be saved into.
overwrite: bool
If False, will throw error when attempting to overwrite an existing file. If set to True, data will be overwritten with a warning.
sep: str
Seperator between each type of tag.
Returns
--------
new_filepath: str
Location of the reformatted file.
"""
ignored_labels = [key for key, value in transformed_filepath_dic.items() if value == "nan"]
if new_filepath is None:
split = filepath.split("/")
new_filepath = "/".join(split[:-1] + ["transform_" + split[-1]])
if os.path.exists(new_filepath):
if overwrite:
print(
f"Warning: File {new_filepath} already exists, overwriting existing file with reformatted data. "
)
else:
print(
f"Error: File {new_filepath} already exists, cannot overwrite existing file. Either set overwrite=True or provide a path to a new file."
)
with open(filepath) as lines, open(new_filepath, "w") as new_file:
data, sentence, label = [], [], []
for line in lines:
if len(line) == 0 or line.startswith("-DOCSTART") or line[0] == "\n":
new_file.write(line)
continue
splits = line.split(sep)
label = splits[-1][:-1]
if (
label in ignored_labels
): # Ignore NEs that are marked as "nan" in transformed_filepath_dic
continue
splits[-1] = transformed_filepath_dic[label] + splits[-1][-1]
new_line = sep.join(splits)
new_file.write(new_line)
return new_filepath
def read_from_huggingface(dataset_path):
"""Reads Hugging Face dataset from dataset bank at dataset_path. Data should be in same format as data in TNER dataset bank."""
dataset = load_dataset(dataset_path)
data_instances = dataset.keys()
if isinstance(data_instances, str):
data_instances = [data_instances]
given_words = []
given_labels = []
for instance in data_instances:
instance_dataset = dataset[instance]
given_words.extend(dataset[instance]["tokens"])
given_labels.extend(dataset[instance]["tags"])
return given_words, given_labels
def read_local_data(filepath, sep=" "):
"""Reads local data that is in a format like the CONLL-2003 dataset from a single string filepath or a list of local files."""
# This code is adapted from: https://github.com/kamalkraj/BERT-NER/blob/dev/run_ner.py
lines = open(filepath)
data, sentence, label = [], [], []
for line in lines:
if len(line) == 0 or line.startswith("-DOCSTART") or line[0] == "\n":
if len(sentence) > 0:
data.append((sentence, label))
sentence, label = [], []
continue
splits = line.split(sep)
word = splits[0]
if len(word) > 0 and word[0].isalpha() and word.isupper():
word = word[0] + word[1:].lower()
sentence.append(word)
label.append(splits[-1][:-1])
if len(sentence) > 0:
data.append((sentence, label))
given_words = [d[0] for d in data]
given_labels = [d[1] for d in data]
return given_words, given_labels
def read_ner_data(dataset_path, format):
"""Function that takes in NER data and returns an array of given words and given labels for each example.
Compatible with "conll2003_like" and "from_huggingface" formats.
"""
if format == "conll2003_like":
if isinstance(dataset_path, str):
dataset_path = [dataset_path]
given_words, given_labels = [], []
for data in dataset_path:
words, labels = read_local_data(data, sep=" ")
given_words.extend(words)
given_labels.extend(labels)
elif format == "from_huggingface":
given_words, given_label = read_from_huggingface(dataset_path)
else:
print(
f"Error. format=={format} is not a supported as a NER data format at this time. Supported formats: [conll2003_like, from_huggingface]."
)
return None
ner_df = create_ner_df(given_words, given_labels)
return ner_df
def create_ner_df(given_words, given_labels):
"""Transforms NER given_words and given_labels into a format required by Studio."""
sentences = list(map(get_sentence, given_words))
sentences, mask = filter_sentence(sentences)
given_words = [words for m, words in zip(mask, given_words) if m]
given_labels = [labels for m, labels in zip(mask, given_labels) if m]
unique_labels = [list(np.unique(label)) for label in given_labels]
labels = [",".join(str(k) for k in label) for label in unique_labels]
id = range(len(labels))
ner_df = pd.DataFrame([id, sentences, given_labels, labels]).transpose()
ner_df.columns = ["id", "tokens", "tags", "tag_sets"]
ner_df["tags"] = ner_df["tags"].astype(str)
return ner_df
def get_suggested_labels(cleanlab_cols):
"""Returns suggested labels as a list of labels for each example provided by cleanlab_cols."""
suggested_labels = cleanlab_cols["suggested_label"]
suggested_labels = [label.split(",") for label in suggested_labels]
return suggested_labels
def get_issue_df(ner_df, cleanlab_cols):
"""Returns NER results in a dataframe containing original labels, cleanlab_cols and "issue_details" column which summarizes NER issues
found in each example.
"""
cleanlab_issue_col_names = [
"cleanlab_row_ID",
"is_label_issue",
"label_issue_score",
"is_well_labeled",
"is_near_duplicate",
"near_duplicate_score",
"near_duplicate_cluster_id",
"is_ambiguous",
"ambiguous_score",
]
df_suggested_given = ner_df[["id", "tag_sets"]].merge(
cleanlab_cols[["suggested_label"]], how="right", left_index=True, right_index=True
)
suggested_labels = df_suggested_given["suggested_label"]
suggested_labels = [label.split(",") for label in suggested_labels]
given_labels = [tag.split(",") for tag in df_suggested_given["tag_sets"]]
suggested_but_not_given = [
np.setdiff1d(suggested, given).tolist()
for given, suggested in zip(given_labels, suggested_labels)
]
given_but_not_suggested = [
np.setdiff1d(given, suggested).tolist()
for given, suggested in zip(given_labels, suggested_labels)
]
ne_issue_column = []
for suggested, given, is_issue in zip(
suggested_but_not_given, given_but_not_suggested, cleanlab_cols["is_label_issue"]
):
issue_detail = ""
if not is_issue:
issue_detail = np.nan
elif len(suggested) == 0 and len(given) == 0:
issue_detail = "This sentence contains a label issue with one or more named entities."
else:
issue_detail = ""
if len(suggested) > 0:
issue_detail += (
"Possible named entitiy tags that are missing: " + ",".join(suggested) + ". "
)
if len(given) > 0:
issue_detail += (
"Possible named entities tags that are incorrectly present: "
+ ",".join(given)
+ ". "
)
ne_issue_column.append(issue_detail)
issue_df = cleanlab_cols[cleanlab_issue_col_names]
issue_df.insert(1, "issue_details", ne_issue_column)
return issue_df
Load Data into Cleanlab Studio
Once we have our data in an acceptable format, we can use the helper function upload_ner_data() to upload our dataset into Cleanlab Studio. The function returns the dataset_id which we will need for the following step to start a project. It takes in the following arguments:
- api_key: Your Cleanlab Studio API key which you can find by going to studio.cleanlab.ai/upload, clicking “Upload via Python API”, and copying the API key there
- dataset_path: The location of the dataset you want to run Named Entity Recognition on. If this data is in “from_huggingface” data format then
dataset_pathshould be the Hugging Face hub data path. If data is in “conll2003_like” format, then the path can be a single local file or list of local file paths. - data_format: Expected format of NER data to be read in. Acceptable formats are described in the Download and Prepare Raw Dataset section above.
- dataset_name (optional): Name for your dataset in Cleanlab Studio.
def upload_ner_data(
api_key,
dataset_path,
data_format,
dataset_name="NER_tutorial_example_dataset",
):
"""Uploads dataset from dataset_path to Cleanlab Studio and returns the dataset_id.
Parameters
----------
api_key: str
Cleanlab Studio API key.
dataset_path: str, list
Location of dataset (either local path or name of the dataset in hugging face).
data_format: str
Expected format of NER data to be read in. Acceptable formats: {"conll2003_like", "from_huggingface"}.
dataset_name: str
Name for your dataset in Cleanlab Studio.
Returns
--------
dataset_id: str
Cleanlab Studio ID of the uploaded dataset.
"""
# Start Cleanlab Studio
studio = Studio(api_key)
# Upload Data to Studio
ner_df = read_ner_data(dataset_path, format=data_format)
display(ner_df.head())
dataset_id = studio.upload_dataset(ner_df, dataset_name=dataset_name)
return dataset_id
For our example, let’s transform the beginning and continuation named entity tags into single tags has_[entity] because we are interested in detecting annotation errors at the entity-level not with respect to beginning vs. continuation subcategories. Let’s also group all miscellaneous entities together under has_other using reformat_ner_data() as described in Reformatting Named Entity Recognition Data.
In your data, similarly group the entities that you are not interested in distinguishing between to avoid seeing a bunch of Cleanlab outputs related to trivial distinctions between entities within the same group. We then pass in our API key and transformed filepaths into upload_ner_data() to upload the dataset to Cleanlab Studio.
filepaths = [
"./data/train.txt",
"./data/valid.txt",
"./data/test.txt",
] # test with multiple filepaths
transform_data_entity_map = {
"O": "has_other",
"B-MISC": "has_other",
"I-MISC": "has_other",
"B-PER": "has_person",
"I-PER": "has_person",
"B-ORG": "has_organization",
"I-ORG": "has_organization",
"B-LOC": "has_location",
"I-LOC": "has_location",
}
dataset_path = [
reformat_ner_data(filepath, transform_data_entity_map, overwrite=True) for filepath in filepaths
]
data_format = "conll2003_like" # Change this to from_huggingface if dataset_path points to a huggingface dataset.
print(f"data_format: {data_format}\ndataset_path: {dataset_path}")
dataset_id = upload_ner_data(api_key=API_KEY, dataset_path=dataset_path, data_format=data_format)
print(f"Uploaded dataset_id: {dataset_id}")
| id | tokens | tags | tag_sets | |
|---|---|---|---|---|
| 0 | 0 | Eu rejects German call to boycott British lamb. | ['has_organization', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other'] | has_organization,has_other |
| 1 | 1 | Peter Blackburn | ['has_person', 'has_person'] | has_person |
| 2 | 2 | Brussels 1996-08-22 | ['has_location', 'has_other'] | has_location,has_other |
| 3 | 3 | The European Commission said on Thursday it disagreed with German advice to consumers to shun British lamb until scientists determine whether mad cow disease can be transmitted to sheep. | ['has_other', 'has_organization', 'has_organization', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other'] | has_organization,has_other |
| 4 | 4 | Germany's representative to the European Union's veterinary committee Werner Zwingmann said on Wednesday consumers should buy sheepmeat from countries other than Britain until the scientific advice was clearer. | ['has_location', 'has_other', 'has_other', 'has_other', 'has_other', 'has_organization', 'has_organization', 'has_other', 'has_other', 'has_other', 'has_person', 'has_person', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_location', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other', 'has_other'] | has_location,has_organization,has_other,has_person |
Train Model to Analyze the CONLL-2003 Data
Once we have our data in an acceptable format, we can launch a Project in Cleanlab Studio. A Project automatically trains ML models that analyze the data for various issues, which takes some time to complete. To launch a Project, use the launch_ner_project() function, which takes in the following arguments:
- api_key: Your Cleanlab Studio API key which you can find by going to studio.cleanlab.ai/upload, clicking “Upload via Python API”, and copying the API key there.
- dataset_id: Cleanlab Studio ID of a dataset uploaded using
upload_ner_data() - project_name (optional): Name for resulting project.
- model_type (optional): Type of model to train (either ‘fast’ or ‘regular’). Cleanlab Studio’s analysis of your dataset is based on training a ML model. A fast model may train quicker but give inferior results. If argument is not specified, a regular model is trained to return the best results.
def launch_ner_project(
api_key, dataset_id, project_name="NER_tutorial_example_project", model_type="regular"
):
"""Creates project and begins training in Cleanlab Studio with dataset provided in dataset_id.
Parameters
----------
api_key: str
Cleanlab Studio API key.
dataset_id: str
Cleanlab Studio ID of an uploaded dataset.
project_name: str
Name for project in Cleanlab Studio. If no name is provided, the project is titled "NER_tutorial_example_project".
model_type: str
Type of model to train (fast or regular).
Returns
--------
project_id: str
Cleanlab Studio ID of created project.
"""
# Start Cleanlab Studio
studio = Studio(api_key)
project_id = studio.create_project(
dataset_id,
project_name=project_name,
modality="text",
task_type="multi-label",
model_type=model_type,
label_column="tag_sets",
text_column="tokens",
)
return project_id
project_id = launch_ner_project(API_KEY, dataset_id)
print(f"Project successfully created and training has begun! project_id: {project_id}")
Once the Project has been launched successfully and you see your project_id you can feel free to close this notebook. It will take some time for Cleanlab’s AI to train on your data and analyze it. Come back after training is complete (you’ll get an email) and continue with the notebook to review your results.
You should only execute the above cell once per dataset. After launching the Project, you can poll for its status to programmatically wait until the results are ready for review as done below. You can optionally provide a timeout parameter after which the function will stop waiting even if the project is not ready.
Warning: This next cell may take a long time to execute for big datasets.
studio = Studio(API_KEY)
cleanset_id = studio.get_latest_cleanset_id(project_id)
print(f"cleanset_id: {cleanset_id}")
studio.wait_until_cleanset_ready(cleanset_id)
If your Jupyter notebook has timed out during this process, then you can resume work by re-running the cell (which should return instantly if the project has completed; do not create a new Project).
After notebook timeout: you’d also need to re-run the starting cells up to the Initialize Helper Methods section. Finally, redefine dataset_path and data_format as they are printed above. If you applied any reformatting to your data using reformat_ner_data() you should pass in the updated dataset_path pointing to these reformatted files (or apply the same transformations again to the original data). Do not create a new Project though!
Results of the Data Analysis
Once it is ready, you can optionally go view your Project via the web interface to explore the results interactively.
Identifying and analyzing examples with label issues is essential for improving dataset quality and training reliable NER models. Here we proceed programmatically via the issues_df returned by Cleanlab Studio, which lists automatically detected issues in our entity recognition dataset.
def get_ner_results(api_key, cleanset_id, dataset_path, data_format):
"""Takes in cleanset_id and original data and returns DataFrame of issues detected in the dataset.
The rows of this DataFrame correspond to same ordering of text examples in your original dataset.
Parameters
----------
api_key: str
Cleanlab Studio API key.
cleanset_id: str
Cleanlab Studio ID of completed project.
dataset_path: str
Location of dataset (either local path or name of the dataset in hugging face).
data_format: str
Expected format of NER data to be read in. Acceptable formats: {"conll2003_like", "from_huggingface"}.
Returns
--------
issue_df: pd.DataFrame
Dataframe containing original labels, cleanlab_cols and "issue_details" column which summarizes NER issues found in each example.
"""
# Start Cleanlab Studio
studio = Studio(api_key)
# Get Cleanlab Studio results
cleanlab_cols = studio.download_cleanlab_columns(cleanset_id)
# Create an issue df reporting results for NER using Cleanlab
ner_df = read_ner_data(dataset_path, format=data_format)
issue_df = get_issue_df(ner_df, cleanlab_cols)
# Add original columns for readability
issue_df = ner_df[["id", "tokens", "tags"]].merge(
issue_df, how="right", left_index=True, right_index=True
)
issue_df["tags"] = issue_df["tags"].apply(literal_eval)
return issue_df
We want to review the examples most likely annotated with an incorrect set of named entity tags. To do so, we sort the issue_df DataFrame by label_issue_score (descending). The top resulting examples (with the highest label issue scores) are the ones Cleanlab estimates are most likely mislabeled in the original dataset.
For your own dataset, you should review these examples and consider correcting their labels. You can also see which data suffers from other types of issues by sorting issue_df based on other issue scores (such as the ambiguous_score to see the most confusing examples). Each issue score indicates the severity of the issue, where higher values indicate problems worthy of your attention.
CLEANSET_ID = "<insert your cleanset_id>" # We used the ID of the project trained above
issue_df = get_ner_results(
API_KEY, cleanset_id, dataset_path, "conll2003_like"
) # Alternatively, pass in CLEANSET_ID
issue_df = issue_df.sort_values(by=["label_issue_score"], ascending=False)
issue_df.head()
| id | tokens | tags | cleanlab_row_ID | issue_details | is_label_issue | label_issue_score | is_well_labeled | is_near_duplicate | near_duplicate_score | near_duplicate_cluster_id | is_ambiguous | ambiguous_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9072 | 9072 | Sao Paulo 1996-08-27 | [has_person, has_person, has_other] | 9073 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_person. | True | 0.959718 | False | True | 1.000000 | 859 | False | 0.558464 |
| 13237 | 13237 | Denver 1996-08-29 | [has_person, has_other] | 13238 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_person. | True | 0.958938 | False | False | 0.876708 | <NA> | False | 0.563274 |
| 15769 | 15769 | pakistan | [has_other] | 15770 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_other. | True | 0.958618 | False | True | 0.999996 | 830 | False | 0.726484 |
| 18727 | 18727 | Santiago 1996-12-05 | [has_person, has_other] | 18728 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_person. | True | 0.955143 | False | False | 0.442889 | <NA> | False | 0.483547 |
| 9112 | 9112 | Sao Paulo 1996-08-27 | [has_person, has_person, has_other] | 9113 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_person. | True | 0.954981 | False | True | 1.000000 | 859 | False | 0.557956 |
Lets take a closer look at the most likely label issue, example id=18378 below. We can see the original tokens are “Prince Rupert 1 3” while their original tags are [has_location, has_location, has_other, has_other]. We know this example is an issue since the is_label_issue boolean is true and the example’s label_issue_score is close to 1.
Column issue_details will tell us more details on which tags Cleanlab Studio believes contribute to the example being marked a label issue. Here Cleanlab Studio suggests that possible named entity tags that are incorrectly present in the tokens are has_location while tags that are missing from the tokens are has_organization and has_person. Tokens “Prince Rupert” pretty clearly represent a person however they were originally marked as has_location. With the help of issue_details we can confirm that has_location is incorrectly present in the tags. Instead, it should be replaced with one of the missing tokens: has_person.
issue_df.iloc[0:1]
| id | tokens | tags | cleanlab_row_ID | issue_details | is_label_issue | label_issue_score | is_well_labeled | is_near_duplicate | near_duplicate_score | near_duplicate_cluster_id | is_ambiguous | ambiguous_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9072 | 9072 | Sao Paulo 1996-08-27 | [has_person, has_person, has_other] | 9073 | Possible named entitiy tags that are missing: has_location. Possible named entities tags that are incorrectly present: has_person. | True | 0.959718 | False | True | 1.0 | 859 | False | 0.558464 |
Learn more about the dataset
In addition to using Cleanlab Studio to detect label errors, we can also identify examples that are: well labeled or ambiguous.
This information provides additional insights about our data annotations.
Examples marked as well labeled are accurately annotated (with great confidence according to Cleanlab’s AI) and do not require extra review from human annotators. These instances are indicated by True in the is_well_labeled column. Given the prevalence of label issues in this dataset, auto-marking examples as “well labeled” is exercised with caution, resulting in no instances here.
print(
"Number of well labeled examples in dataset: ",
issue_df[issue_df["is_well_labeled"] == True].shape[0],
)
issue_df[issue_df["is_well_labeled"] == True]
| id | tokens | tags | cleanlab_row_ID | issue_details | is_label_issue | label_issue_score | is_well_labeled | is_near_duplicate | near_duplicate_score | near_duplicate_cluster_id | is_ambiguous | ambiguous_score |
|---|
Cleanlab Studio can also auto-detect which examples are ambiguous. These are examples that seemingly may or may not contain the given tags, it will be hard for your data annotators to decide correctly without precise instructions on how to handle such cases (different annotators may disagree). In this dataset, there are 68 examples judged to be ambiguous. Looking at one of the examples with a high ambiguity score, we can guess that the token “Karlsruhe” is a city in Germany but could also represent a team name or an individual player. Without additional context, annotators may struggle between choosing has_organization, has_location and has_person.
Despite being ambiguous, this example is not considered a label issue. This is a common as ambiguous examples may be correctly labeled but still confusing and worthy of note.
ambiguous_df = issue_df[issue_df["is_ambiguous"] == True]
print("Number of ambiguous examples in dataset: ", ambiguous_df.shape[0])
# sort by ambiguous score (most ambiguous first)
ambiguous_df = ambiguous_df.sort_values(by="ambiguous_score", ascending=False)
ambiguous_df.iloc[[10]]
| id | tokens | tags | cleanlab_row_ID | issue_details | is_label_issue | label_issue_score | is_well_labeled | is_near_duplicate | near_duplicate_score | near_duplicate_cluster_id | is_ambiguous | ambiguous_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14586 | 14586 | Karlsruhe won the August 20 match 3-1 thanks to two late goals. | [has_organization, has_other, has_other, has_other, has_other, has_other, has_other, has_other, has_other, has_other, has_other, has_other, has_other] | 14587 | NaN | False | 0.446649 | False | False | 0.1052 | <NA> | True | 0.999984 |