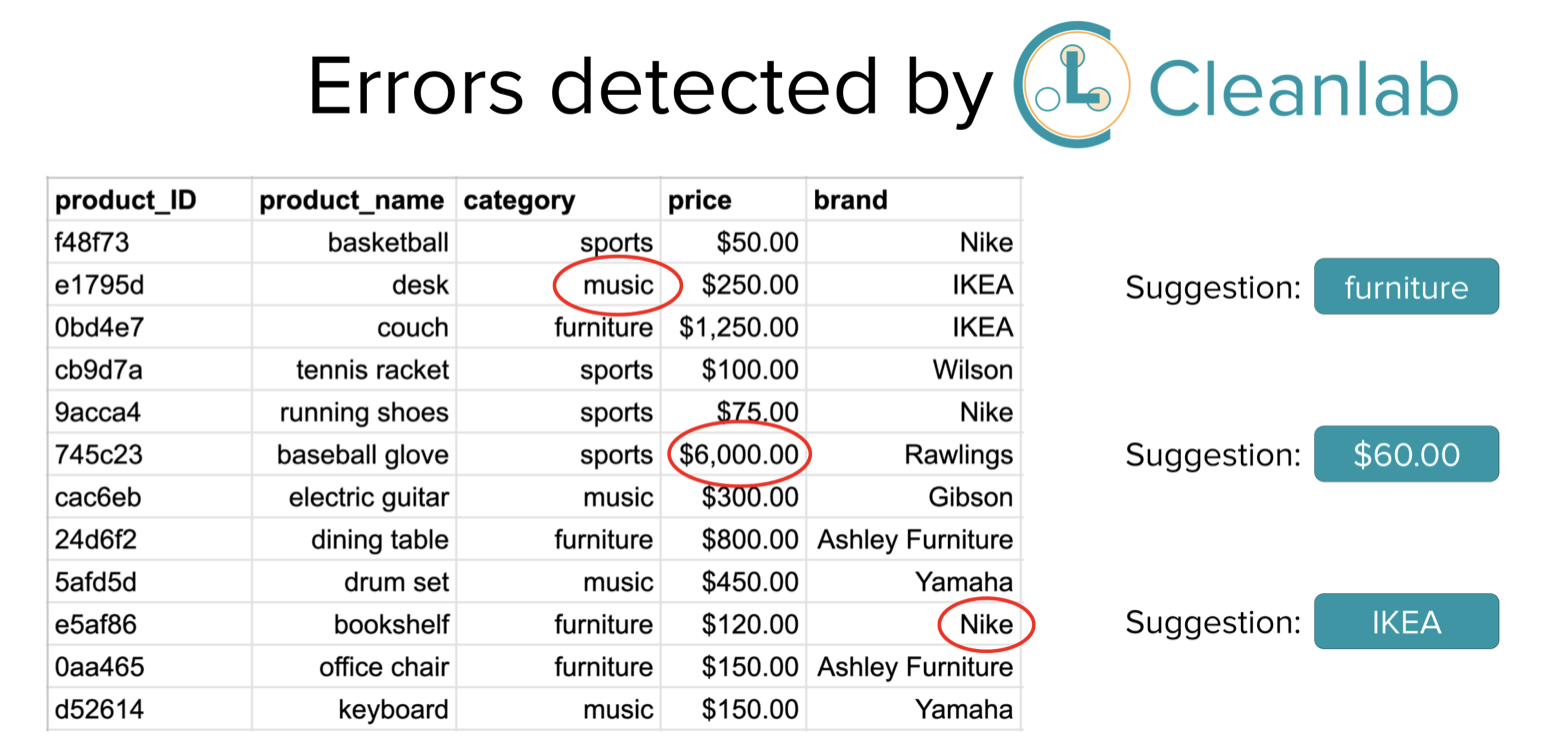
Detect Data Entry Errors (and Impute Missing Values) in any Tabular Dataset
This is the recommended tutorial for programmatically identifying erroneous values (or imputing missing values) in a structured dataset via the Cleanlab Studio Python API. Cleanlab’s AI automatically detects entries in any data table (e.g. CSV/Excel file or Database) that are likely incorrect, perhaps due to: data entry or measurement error (e.g. sensor noise).
Simply provide any data table (including columns that are: text, numeric, or categorical — even with missing values), and state-of-the-art ML models will be trained to score the quality of each datapoint (row) and flag any entry (cell value) that is likely erroneous. These same ML models can produce predictions to accurately impute missing entries in the table.
Unlike traditional data validation or data quality tools, Cleanlab is not simply based on manual rules (e.g. ‘this column only contains positive values in a certain range’). Instead Cleanlab uses AI that accounts for all of the available information in your dataset to flag suspicious entries which may be erroneous. Thus Cleanlab can automatically detect not only atypical values in a column (like traditional data quality tools), but also atypical combinations of values across columns (e.g. multiple values that are jointly incompatible).
Install and import dependencies
You can use pip to install all other packages required for this tutorial as follows:
%pip install cleanlab-studio
import numpy as np
import pandas as pd
import os
import random
from IPython.display import display, Markdown
pd.set_option("display.max_colwidth", None)
Fetch and view dataset
This tutorial considers a structured dataset of medical records. To fetch this dataset, make sure you have wget installed.
wget -nc https://cleanlab-public.s3.amazonaws.com/Datasets/data_entry.csv -P data
BASE_PATH = os.getcwd()
dataset_path = os.path.join(BASE_PATH, "data/")
data = pd.read_csv(os.path.join(dataset_path, 'data_entry.csv'))
data.head()
| patient_id | diagnosis | medication | dosage | note | visiting_hours | invoice | |
|---|---|---|---|---|---|---|---|
| 0 | 90235 | Bronchitis | Dextromethorphan | 10mg | Adjusting dosage of Dextromethorphan for Bronchitis. | 2.0 | 40.0 |
| 1 | 34227 | Pneumonia | Amoxicillin | 500mg | Reviewing patient's response to Amoxicillin therapy for Pneumonia. | 35.0 | 300.0 |
| 2 | 21253 | Hypertension | Hydrochlorothiazide | 25mg | Recommending additional tests to monitor efficacy of Hydrochlorothiazide for Hypertension. | 3.0 | 300.0 |
| 3 | 44136 | Pneumonia | Levofloxacin | 10 IU | Advised patient on importance of adherence to Levofloxacin regimen for Pneumonia. | 5.0 | 300.0 |
| 4 | 76265 | Bronchitis | Guaifenesin | 200mg | Patient treated for Bronchitis using Guaifenesin. | 4.0 | 80.0 |
We can check if the dataset is complete or if there are missing entries.
missing_data = data[data.isnull().any(axis=1)]
missing_data
| patient_id | diagnosis | medication | dosage | note | visiting_hours | invoice | |
|---|---|---|---|---|---|---|---|
| 102 | 10322 | Hypertension | Losartan | NaN | Patient presenting with Hypertension symptoms despite Losartan therapy. | 4.0 | 400.0 |
| 129 | 53909 | Pneumonia | Azithromycin | 250mg | NaN | 5.0 | 300.0 |
| 256 | 68289 | Bronchitis | Guaifenesin | 200mg | Patient responding well to Guaifenesin for Bronchitis. | 5.0 | NaN |
| 397 | 81520 | Diabetes mellitus | Insulin | 10 IU | Considering alternative treatments for Diabetes mellitus due to patient's intolerance to Insulin. | 2.0 | NaN |
| 473 | 61117 | Diabetes mellitus | Insulin | 10 IU | Instructed patient on proper administration of Insulin for Diabetes mellitus. | NaN | 80.0 |
| 655 | 64952 | Influenza | Rimantadine | 100mg | Patient presenting with Influenza symptoms despite Rimantadine therapy. | NaN | 80.0 |
| 816 | 15877 | Bronchitis | NaN | 10mg | Scheduled follow-up appointment to evaluate Bronchitis. | 1.0 | 20.0 |
| 966 | 49846 | Bronchitis | NaN | 10mg | NaN | 1.0 | 20.0 |
Cleanlab Studio automatically deals with missing values, so we don’t have to specially handle them.
Load data into Cleanlab Studio
Our data analysis starts by loading the data, which may take a while for big datasets. First use your API key to instantiate a Studio object.
from cleanlab_studio import Studio
# You can find your Cleanlab Studio API key by going to studio.cleanlab.ai/upload,
# clicking "Upload via Python API", and copying the API key there
API_KEY = "<insert your API key>"
# Initialize studio object
studio = Studio(API_KEY)
A unique ID column allows us to better track and visualize results. If such a column is not present in your dataset, you can uncomment the following code to create one.
Here we’ll use the ‘patient_id’ column which has unique values for all entries in our dataset.
# Alternatively you can create an ID column of sequential integers
# identifier_col = 'unique_id'
# data[identifier_col] = range(0, len(data))
identifier_col = 'patient_id' # unique ID column
data[identifier_col].is_unique
When loading data into Cleanlab Studio, the application will automatically infer the schema of your dataset. However in order to analyze specific numeric columns later on, we’ll override the default schema here to explicitly declare these as numeric columns. You don’t need to do this for all numeric columns, only certain columns you wish to specifically analyze/predict within a later Cleanlab Studio Project.
dataset_id = studio.upload_dataset(
data,
dataset_name="tabular-data-entry",
schema_overrides=[
{"name": "visiting_hours", "column_type": "float"},
{"name": "invoice", "column_type": "float"},
],
)
print(f"Dataset ID: {dataset_id}")
After it’s loaded, we can use the dataset’s id to create a Project in Cleanlab Studio, which automatically trains ML models to provide AI-based analysis of your dataset.
Launch projects
To find erroneous values in this dataset, we launch Cleanlab Studio project for each numeric/categorical column which may contain errors. Each project uses ML to analyze a particular column of the dataset by treating that column as the label (dependent variable), and the rest as predictive features (independent variables). The predictions for each entry in the label column are thus based on all of the other available information in the same row, and can be used to impute missing entries and detect erroneous entries.
We first define helper functions to determine the type of each column and launch Cleanlab Studio projects for a column (based on its determined type).
Optional: Helper functions to determine the type of a column, in terms of the Cleanlab project to find erroneous values in it.
def is_categorical_column(col, unique_threshold = 0.05, min_na_values = 100):
"""
Check if a column is categorical and can be used as the labels for a Cleanlab Studio's classification project.
Parameters:
col (pd.Series or pd.DataFrame): Values in this column (extracted from the original DataFrame).
unique_threshold (float): Threshold to check uniqueness of categories in the column. If the ratio of (unique categories in the column / total number of data samples) is too high, return False.
min_na_values (int): Minimum number of non-null values in this column in order to train a reliable AI model (must be above this to return True).
Returns:
bool: Whether the column is categorical and can be used as label in Cleanlab Studio project.
"""
num_unique_vals = col.nunique()
# Check if column have all identical values
if num_unique_vals == 1:
return False
# Check if column can be considered to be categorical
unique_ratio = num_unique_vals / len(col)
if unique_ratio > unique_threshold:
return False
# Check the minimum class count for any class to be at least 5
if any(col.value_counts() < 5):
return False
# Check if too many NaN values to be worth training a model to predict
if sum(col.notna()) < min_na_values:
return False
if len(col) < 101 and sum(col.isna()) > 0:
return False
return True
def is_numeric_column(col, min_na_values = 100):
"""
Check if a column is numeric and if it can be used as the labels for a Cleanlab Studio regression project.
Parameters:
col (pd.Series or pd.DataFrame): Values in this column (extracted from the original DataFrame).
min_na_values (int): Minimum number of non-null values in this column in order to train a reliable AI model (must be above this to return True).
Returns:
bool: Whether the column is numeric and can be used as label in Cleanlab Studio project.
"""
if col.dtype not in (float, int):
return False
# Check if too many NaN values to be worth training a model to predict
if sum(col.notna()) < min_na_values:
return False
if len(col) < 101 and sum(col.isna()) > 0:
return False
return True
def filter_column_types(data, identifier_col, columns_to_exclude = []):
"""
Determine each column's type: categorical or numerical, to determine which type of Cleanlab Studio project to run for this column.
Parameters:
data (pd.DataFrame): Original dataset.
identifier_col (str): Unique identifier column (will not be considered).
columns_to_exclude (list[str]): List of names of columns to not consider.
Returns:
categorical_columns (list): List of names of columns identified as categorical
numeric_columns (list): List of names of columns identified as numeric
skip_columns (list): List of names of columns that are neither categorical nor numerical (e.g. contain arbitrary text or strings).
"""
categorical_columns = []
numeric_columns = []
skip_columns = []
all_columns = data.columns.to_list()
# List of columns to analyse
columns_to_analyze = [column for column in all_columns if column not in columns_to_exclude]
# Remove identifier_col as it doesn't require any analysis
columns_to_analyze.remove(identifier_col)
for col_name in columns_to_analyze:
col = data[col_name]
if is_categorical_column(col):
categorical_columns.append(col_name)
elif is_numeric_column(col):
numeric_columns.append(col_name)
else:
skip_columns.append(col_name)
if len(categorical_columns + numeric_columns) == 0:
print("No columns to find errors in.")
return categorical_columns, numeric_columns, skip_columns
# Helper function to launch Cleanlab project to detect errors in a column
def create_column_project(dataset_id, project_name, input_columns, audit_column, task_type, model_type="regular"):
"""
Launch Cleanlab Studio project based on the type of the column to be audited.
Parameters:
dataset_id (str): Cleanlab Studio dataset ID.
project_name (str): Name of the Cleanlab Studio project.
input_columns (list[str]): List of names of columns to use as predictive features for training Cleanlab AI model to analyze the `audit_column`.
audit_column (str): Name of the column to audit i.e. use as label for training Cleanlab AI model.
task_type (str): Type of ML task - 'multi-class' for classification or 'regression' for regression.
model_type (str): Type of ML model - 'regular' or 'fast' mode.
Returns:
project_id (str): Unique ID of this Cleanlab Studio project that can be used to programmatically fetch results.
"""
predictive_columns = input_columns.copy()
# Remove audit_column from the list of input predictive features
predictive_columns.remove(audit_column)
project_id = studio.create_project(
dataset_id=dataset_id,
project_name=f"{project_name}-{audit_column}",
modality="tabular",
task_type=task_type,
model_type=model_type,
label_column=audit_column,
feature_columns=predictive_columns
)
print(f"Project for auditing '{audit_column}' created and training has begun! project_id: {project_id}")
return project_id
Not all columns may be susceptible to data entry errors. To save time, you can specify columns_to_exclude to skip these columns in the error checks (these columns may still be used as predictive features for inferring whether other columns have erroneous values).
In our dataset, we’ll skip the ‘diagnosis’ column. Note that any identifier column you previously declared for the dataset (in our case ‘patient_id’) will be ignored (neither being checked for errors nor used as a predictive feature).
The following code determines which categorical and numeric columns we will check for errors (i.e. run a project for).
# List of columns to exclude
columns_to_exclude = ['diagnosis']
categorical_columns, numeric_columns, skip_columns = filter_column_types(data, identifier_col, columns_to_exclude)
print("Categorical columns to audit:", categorical_columns, "\nNumeric columns to audit:", numeric_columns, "\nNot categorical or numeric (not audited):", skip_columns)
We only audit numeric or categorical columns for errors (the ‘note’ column in our dataset contains free-form text and is automatically not checked for errors). For each column to audit, an appropriate Cleanlab project is launched (classification project for categorical columns, regression project for numeric columns). Let’s launch these projects:
project_name = "tutorial" # You can change this prefix used to name the per-column projects
model_type = "regular" # You can set this to "fast" to get quicker but less accurate results
# All the columns (except identifier) can be predictive features
input_columns = data.columns.to_list() # You can reduce this to use less predictive features
input_columns.remove(identifier_col)
project_ids = {} # will store IDs of all the projects for different columns
# Run classification projects for categorical columns
for audit_column in categorical_columns:
project_ids[audit_column] = create_column_project(dataset_id, project_name, input_columns, audit_column, 'multi-class', model_type)
# Run regression projects for numeric columns
for audit_column in numeric_columns:
project_ids[audit_column] = create_column_project(dataset_id, project_name, input_columns, audit_column, 'regression', model_type)
Once all of the projects have been launched successfully and their project_id’s are visible, feel free to close this notebook. It will take time for Cleanlab’s AI to train and analyze your data. Come back after training is complete (you will receive emails) and continue with the notebook to review your results. Each project produces a cleanset (cleaned dataset). For this tutorial, all projects launched here should complete within around 10 minutes.
You should only execute the above cells once! Do not call create_project again.
Before we proceed with the rest of the tutorial, we will wait for all the projects to complete. You can poll for project’s status to programmatically wait until the results are ready for review (this next code cell will take some time):
cleanset_ids = {}
for audit_column in project_ids:
cleanset_ids[audit_column] = studio.get_latest_cleanset_id(project_ids[audit_column])
print(f"Project for auditing '{audit_column}' is running! cleanset_id: {cleanset_ids[audit_column]}")
project_status = studio.wait_until_cleanset_ready(cleanset_ids[audit_column])
Get project results
Each project generates smart metadata about each data point, which is stored as Cleanlab columns we can fetch.
If at any point you want to re-run the remaining parts of this notebook (without creating another project), simply call studio.download_cleanlab_columns(cleanset_id) with the cleanset_id printed from the previous cells.
We define a helper function that fetches Cleanlab generated metadata columns for each project and stores: which entries are estimated to be erroneous, confidence scores for these estimates, and AI model predictions in 3 CSV files (as well as returning corresponding DataFrames). Other helper functions are defined to help inspect/highlight Cleanlab results for each column using these 3 DataFrames/CSVs.
Optional: Helper function to extract and re-arrange data entries detected as possibly erroneous
def extract_and_write_cleanlab_results(data, cleanset_ids, identifier_col):
"""
Fetch Cleanlab columns to store erroneous entries, respective confidence scores, and AI model predictions in 3 different CSVs
Parameters:
data (pd.DataFrame): Original data's DataFrame
cleanset_ids (dict): Dictionary with column name as key, and its respective cleanset_id as value
identifier_col (str): Unique identifier column
Returns:
is_issue_df (pd.DataFrame): DataFrame of booleans, for all audited columns, denoting whether the value is erroneous or not
issue_score_df (pd.DataFrame): DataFrame of confidence scores (0-1), for all audited columns, denoting probability of value being erroneous
prediction_df (pd.DataFrame): DataFrame of AI predictions, for all audited columns, that would be used to impute missing values
"""
# Instantiate DataFrames with identifier column
is_issue_df, issue_score_df, prediction_df = (pd.DataFrame(data[identifier_col]) for _ in range(3))
for audit_column in cleanset_ids:
# Download Cleanlab columns from this column's project
cleanlab_columns_df = studio.download_cleanlab_columns(cleanset_ids[audit_column])
data_cleanlab_df = data.merge(cleanlab_columns_df, left_index=True, right_index=True)
# DataFrame to store True/False flag whether the entry seems erroneous
is_issue_df[audit_column] = data_cleanlab_df['is_label_issue']
# DataFrame to store AI's confidence score on whether the entry is erroneous
issue_score_df[audit_column] = data_cleanlab_df['label_issue_score']
# DataFrame to store predicted values
prediction_df[audit_column] = np.where(data_cleanlab_df["is_label_issue"] | data_cleanlab_df[audit_column].isna(),
data_cleanlab_df["suggested_label"],
data_cleanlab_df[audit_column])
# Save DataFrames to CSV files (comment these lines if you prefer not to)
is_issue_df.to_csv("is_issue.csv", index=False)
issue_score_df.to_csv("issue_score.csv", index=False)
prediction_df.to_csv("imputed_values.csv", index=False)
return is_issue_df, issue_score_df, prediction_df
# Helper function to view the Cleanlab analysis results for a particular column
def inspect_column(audit_column):
"""
Create a DataFrame with original data, column to inspect and its respective Cleanlab columns
Parameters:
audit_column (str): Name of the column to inspect
Returns:
merged_df (pd.DataFrame): DataFrame of original data, inspected column and its respective Cleanlab columns
"""
is_issue_col = is_issue_df[audit_column].rename(f"{audit_column}_is_issue")
issue_score_col = issue_score_df[audit_column].rename(f"{audit_column}_issue_score")
predicted_values_col = predicted_value_df[audit_column].rename(f"{audit_column}_predicted_value")
# Moving the column for ease of comparison
data_columns = data.columns.to_list()
data_columns.remove(audit_column)
data_columns.append(audit_column)
merged_df = pd.concat([data[data_columns], is_issue_col, issue_score_col, predicted_values_col], axis=1)
return merged_df
# Helper function to highlight particular values for ease of visualization
def apply_highlight(row, col, mask):
'''
Highlight the (row,col) entry if mask[row,col] is True
Parameters:
mask (pd.DataFrame): Contains boolean indicating whether the entry is to be highlighted
Returns:
CSS-style (str): Style to apply to (row, col) entry
'''
# Check if the mask is True for this particular entry
if mask.at[row.name, col]:
return 'background-color: red'
else:
return ''
# Helper function to highlight entries in Excel basis a boolean table
def highlight_errors_in_excel(excel_path, issue_csv_path):
"""
Highlights entries in an Excel file based on boolean values in a DataFrame.
Parameters:
excel_path (path in str): Path of Excel file to manipulate
issue_csv_path (path in str): Path of `is_issue.csv` containing boolean table
Returns:
None: Save the modified Excel file
"""
# Export original data to Excel format
data.to_excel(excel_path, index=False)
# Read Excel data and is_issue.csv
excel_workbook = load_workbook(excel_path)
excel_worksheet = excel_workbook.active
is_issue_df = pd.read_csv(issue_csv_path)
# Columns to consider for highlighting
# is_issue_df contains the columns we audited
columns_to_highlight = is_issue_df.columns.to_list() # You can reduce this to a list of specific columns
columns_to_highlight.remove(identifier_col)
# Define pattern to apply to a cell
yellow_fill = PatternFill(start_color='FFFF00', end_color='FFFF00', fill_type='solid')
# Columns with their respective indice in Excel
excel_column_with_indice = {cell.value:cell.column for cell in excel_worksheet[1]}
# Iterate over columns_to_highlight
for column in columns_to_highlight:
excel_column_idx = excel_column_with_indice[column]
# Apply highlight if the value in `is_issue_df` is True
for row_idx, value in enumerate(is_issue_df[column]):
excel_row_idx = row_idx + 2 # Excel rows indice starts from 1 and first row contains the column headers itself, hence add 2
if value == True: excel_worksheet.cell(row=excel_row_idx, column=excel_column_idx).fill = yellow_fill
excel_workbook.save(excel_path)
This helper function outputs 3 CSV files:
-
is_issue.csvcontains True/False values specifying whether each entry value in the input dataset is estimated to be likely erroneous (True) or not (False). You can filter based on the True values to determine which subset of data appears suspicious/corrupted in your dataset (inferred based on the other available information in the dataset). -
issue_scores.csvcontains scores between 0 and 1 estimating the likelihood that each entry value is corrupted (higher scores indicate values that appear more suspicious/erroneous). You can sort a column’s values by these scores (in descending order) to prioritize which values to review (i.e. which entries are most suspicious). -
imputed_values.csvcontains a value for each entry predicted by Cleanlab’s AI model. The prediction represents the expected value of this entry based on all of the other information in dataset (in particular this row), and can be used as an imputed value if the original value were missing.
All 3 CSV files returned contain the same number of rows as the original dataset, and the columns that we ran through Cleanlab projects. Each erroneous flag (boolean), suspicion score, and imputed value corresponds to the data entry at that same row/column in your original dataset.
The entries flagged with the highest issue scores are those you should inspect first. After that, consider having your team review the entries that Cleanlab has flagged as seeming erronous (based on boolean is_issue.csv).
# Create the above 3 DataFrames
is_issue_df, issue_score_df, predicted_value_df = extract_and_write_cleanlab_results(data, cleanset_ids, identifier_col)
# Display the 3 CSV files saved to the disk
ls -l i*.csv
We can view the values in each of these DataFrames/CSVs.
is_issue_df.head(3)
| patient_id | medication | dosage | visiting_hours | invoice | |
|---|---|---|---|---|---|
| 0 | 90235 | False | False | False | False |
| 1 | 34227 | False | False | True | False |
| 2 | 21253 | False | False | False | False |
issue_score_df.head(3)
| patient_id | medication | dosage | visiting_hours | invoice | |
|---|---|---|---|---|---|
| 0 | 90235 | 0.106462 | 0.468849 | 0.288734 | 0.032805 |
| 1 | 34227 | 0.495846 | 0.471051 | 1.000000 | 0.124499 |
| 2 | 21253 | 0.105022 | 0.469134 | 0.406527 | 0.039586 |
predicted_value_df.head(3)
| patient_id | medication | dosage | visiting_hours | invoice | |
|---|---|---|---|---|---|
| 0 | 90235 | Dextromethorphan | 10mg | 2.000000 | 40.0 |
| 1 | 34227 | Amoxicillin | 500mg | 5.301408 | 300.0 |
| 2 | 21253 | Hydrochlorothiazide | 25mg | 3.000000 | 300.0 |
Optional: Analyze erroneous entries in a spreadsheet
Tabular data is frequently analyzed in Microsoft Excel for its ease of use. We can make use of the is_issue.csv and the original data to create a new Excel file (.xlsx) with erroneous cells highlighted in yellow.
The file highlighted_data_entry.xlsx is saved to the current working directory and can be opened using a spreadsheet software.
Note:
You have the flexibility to flag additional or fewer entries in your Data Table by setting higher or lower thresholds on the entries in issue_scores.csv, allowing you to create your customized boolean table.
# Library for manipulating Excel table
%pip install openpyxl
# Import necessary methods
from openpyxl import load_workbook
from openpyxl.styles import PatternFill
# Path of output Excel file and is_issue.csv
excel_path = 'highlighted_data_entry.xlsx'
issue_csv_path = 'is_issue.csv' # Boolean table that we generated above
# Save the highlighted Excel data
highlight_errors_in_excel(excel_path, issue_csv_path)
Impute missing values
Before detecting data entry errors, let’s first see how to impute the missing entries in the original dataset using Cleanlab’s predicted value in predicted_value_df. These predictions come from state-of-the-art machine learning models trained (on your dataset) to predict this missing entry based on the other information available for this row.
highlight_imputed_values = True # Change to False if you don't want to highlight imputed values
missing_data_imputed = missing_data.copy()
impute_columns = categorical_columns + numeric_columns # You can reduce this to a list of specific columns
for col in impute_columns:
missing_data_imputed[col] = missing_data_imputed[col].fillna(predicted_value_df[col])
if highlight_imputed_values:
# Boolean mask indicating whether the entry is missing or not
mask_impute = missing_data.isna()
# Apply highlight style
missing_data_imputed = missing_data_imputed.style.apply(lambda x: [apply_highlight(x, col, mask_impute) for col in x.index], axis=1, subset=impute_columns)
missing_data_imputed
| patient_id | diagnosis | medication | dosage | note | visiting_hours | invoice | |
|---|---|---|---|---|---|---|---|
| 102 | 10322 | Hypertension | Losartan | 50mg | Patient presenting with Hypertension symptoms despite Losartan therapy. | 4.000000 | 400.000000 |
| 129 | 53909 | Pneumonia | Azithromycin | 250mg | nan | 5.000000 | 300.000000 |
| 256 | 68289 | Bronchitis | Guaifenesin | 200mg | Patient responding well to Guaifenesin for Bronchitis. | 5.000000 | 202.368744 |
| 397 | 81520 | Diabetes mellitus | Insulin | 10 IU | Considering alternative treatments for Diabetes mellitus due to patient's intolerance to Insulin. | 2.000000 | 174.182175 |
| 473 | 61117 | Diabetes mellitus | Insulin | 10 IU | Instructed patient on proper administration of Insulin for Diabetes mellitus. | 1.003176 | 80.000000 |
| 655 | 64952 | Influenza | Rimantadine | 100mg | Patient presenting with Influenza symptoms despite Rimantadine therapy. | 2.114235 | 80.000000 |
| 816 | 15877 | Bronchitis | Dextromethorphan | 10mg | Scheduled follow-up appointment to evaluate Bronchitis. | 1.000000 | 20.000000 |
| 966 | 49846 | Bronchitis | Dextromethorphan | 10mg | nan | 1.000000 | 20.000000 |
Review the data entry error audit for each column
Let’s take a closer look at the ‘medication’ column. Here we obtain a merged DataFrame using the inspect_column helper function defined above, then filter for the data points where is_issue is True and sort the values by issue_score to view the top most suspicious entries detected in this column.
audit_column = 'medication'
medication_df = inspect_column(audit_column)
medication_issues = medication_df[medication_df[f"{audit_column}_is_issue"] == True].sort_values(f"{audit_column}_issue_score", ascending=False)
medication_issues.head()
| patient_id | diagnosis | dosage | note | visiting_hours | invoice | medication | medication_is_issue | medication_issue_score | medication_predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 484 | 73051 | Influenza | 10mg | Patient treated for Influenza using Rimantadine. | 1.0 | 40.0 | Rimantadine | True | 0.920749 | Zanamivir |
| 771 | 11433 | Pneumonia | 500mg | Educated patient on potential side effects of Amoxicillin for Pneumonia. | 2.0 | 120.0 | Insulin | True | 0.709063 | Amoxicillin |
Per drugs.com, Rimantadine is typically prescribed at a dosage of 100mg, not 10mg which is the dosage for Zanamavir (predicted value). Also, Insulin is the medication for Diabetes mellitus, not Pneumonia. Such entries can be referred to subject matter expert for review.
Next let’s check the visiting_hours column and similarly view the top most suspicious values detected in this column.
audit_column = 'visiting_hours'
visiting_hours_df = inspect_column(audit_column)
visiting_hours_issues = visiting_hours_df[visiting_hours_df[f"{audit_column}_is_issue"] == True].sort_values(f"{audit_column}_issue_score", ascending=False)
visiting_hours_issues.head()
| patient_id | diagnosis | medication | dosage | note | invoice | visiting_hours | visiting_hours_is_issue | visiting_hours_issue_score | visiting_hours_predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34227 | Pneumonia | Amoxicillin | 500mg | Reviewing patient's response to Amoxicillin therapy for Pneumonia. | 300.0 | 35.0 | True | 1.0 | 5.301408 |
| 325 | 52005 | Pneumonia | Levofloxacin | 500mg | Patient exhibiting adverse reaction to Levofloxacin prescribed for Pneumonia. | 300.0 | 20.0 | True | 1.0 | 5.519128 |
| 639 | 39637 | Hypertension | Hydrochlorothiazide | 10mg | Instructed patient on proper administration of Hydrochlorothiazide for Hypertension. | 500.0 | 65.0 | True | 1.0 | 7.023181 |
| 710 | 19203 | Hypertension | Losartan | 50mg | Noted improvement in patient's condition after administering Losartan for Hypertension. | 200000.0 | 30.0 | True | 1.0 | 5.218635 |
| 855 | 25119 | Influenza | Rimantadine | 100mg | Recommended lifestyle modifications in addition to Rimantadine therapy for Influenza. | 120.0 | 28.0 | True | 1.0 | 3.762323 |
The visiting hours recorded in these entries appears off as patients don’t typically have these sorts of consultations for such long hours. Such entries can be double checked based on domain knowledge and their presence may indicate you should implement regularly scheduled data validation to ensure values fall within known ranges.
We repeat the same steps as above, but this time for the dosage column to check for entries with suspicious values.
audit_column = 'dosage'
dosage_df = inspect_column(audit_column)
dosage_issues = dosage_df[dosage_df[f"{audit_column}_is_issue"] == True].sort_values(f"{audit_column}_issue_score", ascending=False)
dosage_issues.head()
| patient_id | diagnosis | medication | note | visiting_hours | invoice | dosage | dosage_is_issue | dosage_issue_score | dosage_predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 44136 | Pneumonia | Levofloxacin | Advised patient on importance of adherence to Levofloxacin regimen for Pneumonia. | 5.0 | 300.0 | 10 IU | True | 0.532683 | 500mg |
| 639 | 39637 | Hypertension | Hydrochlorothiazide | Instructed patient on proper administration of Hydrochlorothiazide for Hypertension. | 65.0 | 500.0 | 10mg | True | 0.531477 | 25mg |
| 109 | 38965 | Bronchitis | Guaifenesin | Patient diagnosed with Bronchitis. Prescribed Guaifenesin for treatment. | 1.0 | 20.0 | 100mg | True | 0.531120 | 200mg |
| 484 | 73051 | Influenza | Rimantadine | Patient treated for Influenza using Rimantadine. | 1.0 | 40.0 | 10mg | True | 0.530678 | 100mg |
Levofloxacin is prescribed in milligrams (mg) not insulin units (IU), which signals that the dosage value is definitely wrong. Also, the recommended dosage for Rimantadine, as per drugs.com, is 100mg. Zanamivir, another drug for Influenza, is prescribed with 5-10mg dose. So either the recorded dosage or medication seems off here. The same applies to Hydrochlorothiazide which is usually prescribed with 25-100mg dosage for Hypertension, and to Guaifenesin for treating Bronchitis.
For all of these data entries, Cleanlab’s predicted value seems more appropriate.
Finally, we look for data entry errors in the “invoice” column using the same steps.
audit_column = 'invoice'
invoice_df = inspect_column(audit_column)
invoice_issues = invoice_df[invoice_df[f"{audit_column}_is_issue"] == True].sort_values(f"{audit_column}_issue_score", ascending=False)
invoice_issues.head()
| patient_id | diagnosis | medication | dosage | note | visiting_hours | invoice | invoice_is_issue | invoice_issue_score | invoice_predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 710 | 19203 | Hypertension | Losartan | 50mg | Noted improvement in patient's condition after administering Losartan for Hypertension. | 30.0 | 200000.0 | True | 1.0 | 606.011169 |
The invoice value of $200k is too high. Such an entry should be verified from other records to avoid accounting issues later on.
Improve your dataset by correcting erroneous entries
The best way to ensure that you have high quality data is to manually inspect the entries that Cleanlab identified to have issues (i.e. is_issue = True) and correct them. However, that could be time consuming and hence you can also auto-correct your data by replacing the entries that have been flagged as issues with Cleanlab’s predicted values. We recommend reviewing various predicted values from diverse rows to ensure they are reasonable before doing this.
We demonstrate how to automatically obtain an improved dataset below, where autocorrected_dataset will have the exact same rows and columns as your original dataset, but with the entries estimated to be potentially erroneous replaced with values predicted by Cleanlab based on the other information in the dataset.
autocorrected_dataset = data.copy()
fix_columns = categorical_columns + numeric_columns # You can reduce this to a list of specific columns
impute_missing = False # Change this to True if you want to impute the missing values alongside fixing the erroneous entries
mask_condition = {col: (is_issue_df[col] if not impute_missing else is_issue_df[col] | autocorrected_dataset[col].isna()) for col in fix_columns}
for col in fix_columns:
autocorrected_dataset[col] = autocorrected_dataset[col].mask(mask_condition[col], predicted_value_df[col])
autocorrected_dataset.to_csv('autocorrected_dataset.csv', index=False)
Lastly, we view the first 5 samples of the autocorrected dataset highlighting the corrected entries.
samples = 5
highlight_corrected_values = True
autocorrected_dataset_copy = autocorrected_dataset.copy()
if highlight_corrected_values:
autocorrected_dataset_copy = autocorrected_dataset.head(samples).style.apply(lambda x: [apply_highlight(x, col, is_issue_df) for col in x.index], axis=1, subset=impute_columns)
autocorrected_dataset_copy
| patient_id | diagnosis | medication | dosage | note | visiting_hours | invoice | |
|---|---|---|---|---|---|---|---|
| 0 | 90235 | Bronchitis | Dextromethorphan | 10mg | Adjusting dosage of Dextromethorphan for Bronchitis. | 2.000000 | 40.000000 |
| 1 | 34227 | Pneumonia | Amoxicillin | 500mg | Reviewing patient's response to Amoxicillin therapy for Pneumonia. | 5.301408 | 300.000000 |
| 2 | 21253 | Hypertension | Hydrochlorothiazide | 25mg | Recommending additional tests to monitor efficacy of Hydrochlorothiazide for Hypertension. | 3.000000 | 300.000000 |
| 3 | 44136 | Pneumonia | Levofloxacin | 500mg | Advised patient on importance of adherence to Levofloxacin regimen for Pneumonia. | 5.000000 | 300.000000 |
| 4 | 76265 | Bronchitis | Guaifenesin | 200mg | Patient treated for Bronchitis using Guaifenesin. | 4.000000 | 80.000000 |